Overview
Our research focus concerns one of the most central questions in evolutionary biology: what is the origin of biodiversity? We are fundamentally interested in the mechanisms that create and maintain differences among individuals or among populations / species. These differences can be called discontinuities in nature and include ecological, morphological and genetic/genomic discontinuities. We use a combination of field and lab experiments, as well as surveys of molecular genetic variation to test hypotheses on the mechanisms that drive the evolution of these discontinuities. Below are short summaries of some current projects.
Hybridization and Speciation in Lycaeides Butterflies
The Lycaeides species complex is a polytypic group of butterflies (Family Lycaenidae) that is an ideal system for investigating the mechanisms driving differentiation and the evolution of reproductive isolation because this group contains a number of closely-related entities with a history of isolation, secondary contact and hybridization. These butterflies exhibit 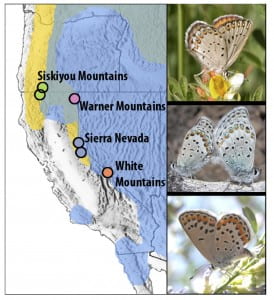 extensive morphological and ecological variation. Across North America, evidence for multiple Pleistocene-aged refugia is evident from phylogeographic patterns of mitochondrial DNA (mtDNA) and nuclear markers. Where these refugial populations meet, gene exchange via hybridization has occurred. In these zones of contact, called suture zones, we find evidence of mtDNA introgression and genomic admixture with multiple potential cases of homoploid hybrid species, which have rarely been demonstrated in animals. Ecological field experiments and population genetic and genomic analyses are used to investigate the evolution and maintenance of morphological and ecological variation, the mechanisms by which reproductive isolation evolves, and the relative contributions of natural selection and other factors that shape hybrid genomes. Collaborators include: Zach Gompert (Utah State), Jim Fordyce (U Tennessee), Matt Forister (U Nevada), Lauren Lucas (Texas State/Utah State), Alex Buerkle (U Wyoming). Read more: gompert14nice13lucas08forister11
extensive morphological and ecological variation. Across North America, evidence for multiple Pleistocene-aged refugia is evident from phylogeographic patterns of mitochondrial DNA (mtDNA) and nuclear markers. Where these refugial populations meet, gene exchange via hybridization has occurred. In these zones of contact, called suture zones, we find evidence of mtDNA introgression and genomic admixture with multiple potential cases of homoploid hybrid species, which have rarely been demonstrated in animals. Ecological field experiments and population genetic and genomic analyses are used to investigate the evolution and maintenance of morphological and ecological variation, the mechanisms by which reproductive isolation evolves, and the relative contributions of natural selection and other factors that shape hybrid genomes. Collaborators include: Zach Gompert (Utah State), Jim Fordyce (U Tennessee), Matt Forister (U Nevada), Lauren Lucas (Texas State/Utah State), Alex Buerkle (U Wyoming). Read more: gompert14nice13lucas08forister11
Comparative Phylogeography of Karst- and Spring-associated Aquatic Organisms
 The Edwards Aquifer, which underlies much of south central Texas, is one of the most diverse aquifer systems on Earth, with a substantial portion of that biodiversity still to be described. This aquifer is also the primary water source for human populations, including the cities of San Antonio and Austin and well over 2 million people. Many of the organisms that are endemic to the springs and deeper parts of the aquifer are listed as threatened or endangered. To inform management plans and to understand the (underground) connectivity of populations utilizing the aquifer, we have conducted phytogeographic surveys or several endemic groups. These studies are designed to quantify the geographic patterns of population genetic variation and patterns of gene flow, and test hypotheses about how these organisms use the underground aquifer connections. The list includes Riffle beetles of the genus Heterelmis, Stygobromus amphipods, Stygoparnus Dryopid beetles, and others. Currently, this project is headed by Ph.D. student Will Coleman (Co-advised with Dr. Benjamin Schwartz) who is generating population genomics data to continue and extend these investigations. In addition, ongoing work with Eurycea salamanders is aimed at defining the boundaries of lineages and quantifying the patterns of gene flow, both historical and contemporary. Work on the salamanders is a collaboration with Pete Diaz (USFWS), Paul Crump (TPWD) and Ruben Tovar (UT Austin). Read more: lucas09, lucas09b, ethridge13, Lucas_et_al-2016-Freshwater_Biology, Nice2021_Article_GeographicPatternsOfGenomicVar.
The Edwards Aquifer, which underlies much of south central Texas, is one of the most diverse aquifer systems on Earth, with a substantial portion of that biodiversity still to be described. This aquifer is also the primary water source for human populations, including the cities of San Antonio and Austin and well over 2 million people. Many of the organisms that are endemic to the springs and deeper parts of the aquifer are listed as threatened or endangered. To inform management plans and to understand the (underground) connectivity of populations utilizing the aquifer, we have conducted phytogeographic surveys or several endemic groups. These studies are designed to quantify the geographic patterns of population genetic variation and patterns of gene flow, and test hypotheses about how these organisms use the underground aquifer connections. The list includes Riffle beetles of the genus Heterelmis, Stygobromus amphipods, Stygoparnus Dryopid beetles, and others. Currently, this project is headed by Ph.D. student Will Coleman (Co-advised with Dr. Benjamin Schwartz) who is generating population genomics data to continue and extend these investigations. In addition, ongoing work with Eurycea salamanders is aimed at defining the boundaries of lineages and quantifying the patterns of gene flow, both historical and contemporary. Work on the salamanders is a collaboration with Pete Diaz (USFWS), Paul Crump (TPWD) and Ruben Tovar (UT Austin). Read more: lucas09, lucas09b, ethridge13, Lucas_et_al-2016-Freshwater_Biology, Nice2021_Article_GeographicPatternsOfGenomicVar.
Ecology and Evolution of Pipevine Swallowtails
Pipevine Swallowtails, Battus philenor, are at the center of a mimicry complex in North America. These butterflies are specialists on pipevine plants (Aristolochia sp.) and sequester aristolochic acids which larvae obtain by eating the plants. These toxic alkaloids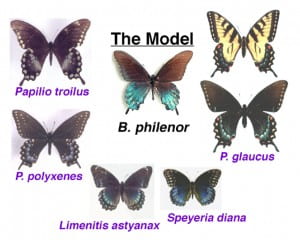 protect caterpillars and adults from predation. Current research on the pipevines is focused on understanding geographical variation in plant toxicity and larval sequestration ability and consequences in a tri-trophic context (plants – caterpillars – natural enemies). We are also interested in life history variation, including clutch size and diapause dynamics, the adaptive significance of larval color variation and the phylogeographic history of this species in North America. Ph.D. student Kate Bell is in charge of this project. Most of this research has been done in collaboration with Jim Fordyce at the University of Tennessee, Knoxville. Read more: dimarco12fordyce08nice06
protect caterpillars and adults from predation. Current research on the pipevines is focused on understanding geographical variation in plant toxicity and larval sequestration ability and consequences in a tri-trophic context (plants – caterpillars – natural enemies). We are also interested in life history variation, including clutch size and diapause dynamics, the adaptive significance of larval color variation and the phylogeographic history of this species in North America. Ph.D. student Kate Bell is in charge of this project. Most of this research has been done in collaboration with Jim Fordyce at the University of Tennessee, Knoxville. Read more: dimarco12fordyce08nice06
The Evolutionary and Ecological Consequences of Aggregative Feeding
Hackberry butterflies or Emperor butterflies (Asterocampa sp. ) are common in southwest United States. The females of both species utilize Hackberry trees (Celtis sp.) as their host plant. Asterocampa celtis (A. celtis) and A. clyton produce different clutch sizes. A. celtis females lay their eggs singly in clutches from 1-20 eggs, while A. clyton females lay their eggs in stacked pyramid shaped clusters of up to 100. Correspondingly, A. celtis larvae feed individually, while A. clyton larvae feed in large, dense aggregations. There are likely advantages and disadvantages to the egg laying strategies of both A. celtis and A. clyton. By laying large clusters of eggs, A. clyton might gain the advantage of gregarious feeding, but might incur the cost of increased parasitism or predation on the more visible, larger clusters of eggs. This alternative strategy and its potential tradeoffs will be the focus of this research using manipulative experiments to compare the benefits of aggregative feeding to caterpillars feeding singly.
